PMC-LLaMA: Towards Building Open-source Language Models for Medicine
PMC-LLaMA: 构建开源医学语言模型的探索
Chaoyi $\mathbf{W}\mathbf{u}^{1,2,* }$ , Weixiong $\mathbf{Lin}^{1,2,* }$ , Xiaoman Zhang1,2, Ya Zhang1,2 Yanfeng Wang1,2, Weidi Xie1,2
Chaoyi $\mathbf{W}\mathbf{u}^{1,2,* }$,Weixiong $\mathbf{Lin}^{1,2,* }$,Xiaoman Zhang1,2,Ya Zhang1,2,Yanfeng Wang1,2,Weidi Xie1,2
1 Cooperative Medianet Innovation Center, Shanghai Jiao Tong University, Shanghai, China 2Shanghai AI Laboratory, Shanghai, China {w tz xxx w cy 02, wx_ lin, xm99sjtu, ya_ zhang, wang yan feng, weidi}@sjtu.edu.cn
1 上海交通大学协同媒体创新中心,中国上海
2 上海人工智能实验室,中国上海
{w tz xxx w cy 02, wx_ lin, xm99sjtu, ya_ zhang, wang yan feng, weidi}@sjtu.edu.cn
Abstract
摘要
Recently, Large Language Models (LLMs) have showcased remarkable capabilities in natural language understanding. While demonstrating proficiency in everyday conversations and question-answering situations, these models frequently struggle in domains that require precision, such as medical applications, due to their lack of domain-specific knowledge. In this paper, we describe the procedure for building a powerful, open-source language model specifically designed for medicine applications, termed as PMC-LLaMA. Our contributions are threefold: (i) we systematically investigate the process of adapting a general-purpose foundation language model towards medical domain, this involves data-centric knowledge injection through the integration of 4.8M biomedical academic papers and 30K medical textbooks, as well as comprehensive fine-tuning for alignment with domain-specific instructions; (ii) we contribute a large-scale, comprehensive dataset for instruction tuning. This dataset encompasses medical question-answering (QA), rationale for reasoning, and conversational dialogues, comprising a total of 202M tokens; (iii) we conduct thorough ablation studies to demonstrate the effectiveness of each proposed component. While evaluating on various public medical question-answering benchmarks, our lightweight PMCLLaMA, which consists of only 13 billion parameters, exhibits superior performance, even surpassing ChatGPT. All models, codes, datasets can be found in https://github.com/ chaoyi-wu/PMC-LLaMA.
近来,大语言模型(LLM)在自然语言理解方面展现出卓越能力。尽管在日常对话和问答场景中表现优异,这些模型由于缺乏领域专业知识,在医疗等需要精准性的领域往往表现不佳。本文阐述了构建专为医疗应用设计的强大开源语言模型PMC-LLaMA的全过程。我们的贡献包含三个方面:(i) 系统研究了通用基础语言模型向医疗领域的适配过程,通过整合480万篇生物医学学术论文和3万本医学教材实现以数据为中心的知识注入,并执行全面的领域指令微调对齐;(ii) 贡献了大规模指令微调数据集,包含医疗问答(QA)、推理依据和对话内容,总计2.02亿token;(iii) 通过详尽的消融实验验证了各模块的有效性。在多个公共医疗问答基准测试中,我们仅含130亿参数的轻量级PMC-LLaMA展现出超越ChatGPT的优异性能。所有模型、代码和数据集详见https://github.com/chaoyi-wu/PMC-LLaMA。
Introduction
引言
The rapid advancement of large language models (LLMs), for example, OpenAI’s ChatGPT (OpenAI 2023b) and GPT4 (OpenAI 2023a) has truly revolutionized the natural language processing research (Nori, King et al. 2023; Singhal et al. 2022), sparking AI applications for numerous daily scenarios. Unfortunately, the training details and model archi tec ture s for the GPT-series remain unclear. The opensource LLMs, e.g., LLaMA-series (Touvron et al. 2023a,b), also show comparable performance with ChatGPT in the general domain. However, though the LLMs demonstrate proficiency in everyday conversations, in medical domain where requires high precision, they often produce seemingly accurate output but lead to incorrect conclusions, which could be highly fatal. We conjecture this is due to their lack of comprehensive medical knowledge.
大语言模型(LLM)的快速发展,例如OpenAI的ChatGPT (OpenAI 2023b)和GPT4 (OpenAI 2023a),彻底改变了自然语言处理研究领域(Nori, King et al. 2023; Singhal et al. 2022),催生了众多日常场景的AI应用。遗憾的是,GPT系列的训练细节和模型架构仍不明确。开源的大语言模型如LLaMA系列(Touvron et al. 2023a,b)在通用领域也展现出与ChatGPT相当的性能。然而,尽管这些大语言模型在日常对话中表现优异,但在需要高精度的医疗领域,它们经常生成看似准确实则错误的结论,这可能造成致命后果。我们推测这是由于它们缺乏全面的医学知识所致。

Figure 1: In the left, we show the general comparison between our PMC-LLaMA with LLaMA-2 and ChatGPT. On the right, we visually show the advantages of our model in model sizes. PMC-LLaMA is much smaller than the others.
图 1: 左侧展示了我们的PMC-LLaMA与LLaMA-2及ChatGPT的总体对比。右侧直观呈现了我们的模型在参数量级上的优势。PMC-LLaMA明显小于其他模型。
Existing works have also explored several ways for adapting general-purpose LLMs towards medicine domain, like Med-Alpaca (Han, Adams et al. 2023), Chat-Doctor (Yunxiang et al. 2023) and MedPALM-2 (Anil, Dai et al. 2023). Among these, MedPALM-2 is the only work successfully outperforming ChatGPT while their training details, for example, training data, model architecture, remain unclear. Thus, systematic investigation on the medical domain adaptation for LLMs still needs to be discussed further especially in open-source community.
现有研究也探索了多种将通用大语言模型适配到医疗领域的方法,例如Med-Alpaca (Han, Adams et al. 2023)、Chat-Doctor (Yunxiang et al. 2023)和MedPALM-2 (Anil, Dai et al. 2023)。其中,MedPALM-2是唯一成功超越ChatGPT的工作,但其训练细节(如训练数据、模型架构)仍不明确。因此,针对大语言模型的医疗领域适配仍需系统性研究,特别是在开源社区中亟待深入探讨。
Our goal is to systematically adapt an open-source general LLM, i.e., LLaMA, towards the medicine domain from the following aspects. First, we adopt data-centric medicalspecific knowledge injection for the language model with a large-scale free text medical corpora. We claim that language models can accumulate enough medical knowledge in this step and build up a better embedding space for domainspecific complex terminologies. Second, augmenting the reasoning capabilities of the proposed model. This empowers the model to link its medical knowledge with provied case information and provide well-justified recommendations. Lastly, enhancing the alignment ability of LLMs. Robust alignment with various instructions facilitates effective zero-shot adaptation to a diverse spectrum of tasks.
我们的目标是从以下几个方面系统性地将开源通用大语言模型(即LLaMA)适配至医疗领域。首先,我们采用以数据为中心的医学专业知识注入方法,通过大规模自由文本医学语料库增强语言模型。我们认为语言模型在此阶段能积累足够的医学知识,并为领域内复杂术语构建更优质的嵌入空间。其次,增强所提模型的推理能力。这使模型能够将其医学知识与提供的病例信息相关联,并给出合理建议。最后,提升大语言模型的对齐能力。与多样化指令的稳健对齐有助于实现针对广泛任务的零样本高效适配。
In conclusion, in this paper we systematically build up an LLM for medicine through data-centric knowledge injection and medical-specific instruction tuning, and release an open-source lightweight medical-specific language model, PMC-LLaMA. Specifically, we first collect a large medical-specific corpus, named MedC-K, consisting of 4.8M biomedical academic papers and 30K textbooks for knowledge injection. We then adopt medical-specific instruction tuning on a new medical knowledge-aware instruction dataset, termed MedC-I, consisting of medical QA, ra- tionale, and conversation with 202M tokens in total. We evaluate PMC-LLaMA on various medical QA benchmarks, surpassing ChatGPT and LLaMA-2 as shown in Fig. 1.
总之,本文通过以数据为中心的知识注入和医学专用指令微调,系统性地构建了一个医学大语言模型,并开源了轻量级医学专用语言模型PMC-LLaMA。具体而言,我们首先收集了名为MedC-K的大型医学专用语料库,包含480万篇生物医学学术论文和3万本教科书用于知识注入。随后基于新型医学知识感知指令数据集MedC-I(包含医学问答、原理阐述及对话,总计2.02亿token)进行医学专用指令微调。如图1所示,PMC-LLaMA在多项医学问答基准测试中表现优于ChatGPT和LLaMA-2。
Related Work
相关工作
Large Language Model. Recently, the great success of large language models (LLM) (OpenAI 2023b,a; Anil, Dai et al. 2023; Du et al. 2021), has garnered significant attention within the field of natural language processing. For example, OpenAI’s strides with ChatGPT and GPT-4 have showcased remarkable capabilities in various tasks, including text generation, language translation, question answering, and more. However, intricate details concerning their training methodologies and weight parameters remain undisclosed. LLaMA (Touvron et al. 2023a) serves as an open-source alternative for the foundational language model, ranging from 7 billion to 65 billion parameters. In light of these advancements, there has been a surge of interest in tailoring language models for specific biomedical domains. Most of these models are prompt-tuned using LLaMA on a small medical corpus, resulting in a deficiency of comprehensive medical knowledge integration.
大语言模型 (Large Language Model)。近期,大语言模型 (LLM) (OpenAI 2023b,a; Anil, Dai et al. 2023; Du et al. 2021) 的巨大成功在自然语言处理领域引起了广泛关注。例如,OpenAI 推出的 ChatGPT 和 GPT-4 在文本生成、语言翻译、问答等多种任务中展现了卓越能力。然而,其训练方法和权重参数等核心细节仍未公开。LLaMA (Touvron et al. 2023a) 作为开源基础语言模型,提供了参数量从 70 亿到 650 亿不等的替代方案。随着这些技术进步,针对生物医学领域定制语言模型的研究兴趣激增。当前多数模型基于 LLaMA 在小规模医学语料上进行提示微调 (prompt-tuning),导致医学知识整合不够全面。
Instruction Tuning. For LLMs to follow natural language instructions and complete real-world tasks, instructiontuning has been widely used for alignment (Ouyang et al. 2022; Peng et al. 2023). This involves fine-tuning the model on a collection of tasks described via instructions, to effectively improve the zero-shot and few-shot generalization abilities of LLMs (Chung et al. 2022; Iyer et al. 2022). Building on the publicly accessible language models, Alpace (Taori et al. 2023) and Vicuna (Chiang, Li, and others. 2023) are proposed, by finetuning on the machine-generated instruction-following samples, showing promising performance. In the medical domain, Chat-Doctor (Yunxiang et al. 2023), and Med-Alpaca (Han, Adams et al. 2023), are instruction-tuned for medical question-answering and dialogue applications. Notably, Med-PaLM (Singhal et al. 2022) represents the pinnacle of LLMs in the medical field, trained with intensive instruction tuning on the strong PaLM model (with 540 billion parameters). However, its code and data remain inaccessible to the public.
指令微调 (Instruction Tuning)。为使大语言模型能遵循自然语言指令并完成现实任务,指令微调被广泛用于对齐 (Ouyang et al. 2022; Peng et al. 2023)。该方法通过在指令描述的任务集合上微调模型,有效提升大语言模型的零样本和少样本泛化能力 (Chung et al. 2022; Iyer et al. 2022)。基于公开可用的语言模型,Alpace (Taori et al. 2023) 和 Vicuna (Chiang, Li, and others. 2023) 通过对机器生成的指令跟随样本进行微调而提出,展现出优异性能。在医疗领域,Chat-Doctor (Yunxiang et al. 2023) 和 Med-Alpaca (Han, Adams et al. 2023) 针对医疗问答和对话应用进行了指令微调。值得注意的是,Med-PaLM (Singhal et al. 2022) 代表了医疗领域大语言模型的巅峰,其基于强大的 PaLM 模型 (5400亿参数) 进行了密集的指令微调训练,但其代码和数据仍未向公众开放。
Medical Foundational Language Model. In addition to instruction tuning, there has been extensive efforts on training foundation model for medicine, for example, BioBert, BioMedGPT, etc. (Lee et al. 2020; Zhang et al. 2023; Luo et al. 2022). However, these models exhibit certain limitations, first, most domain-specific models have been exclusively trained on medical corpora. The lack of exposure to diverse knowledge domains beyond medicine can impede the model’s capability to perform reasoning or context understanding; second, these models are limited in model scale and are predominantly designed to base on BERT, thus imposing restrictions on their utility for a wide array of downstream tasks under zero-shot learning. In this work, we aim to resolve these two limitations by adapting a general LLM toward medicine with knowledge injection, followed by medical-specific instruction tuning.
医疗基础语言模型。除了指令微调外,医学领域的基础模型训练也取得了广泛进展,例如 BioBert、BioMedGPT 等 (Lee et al. 2020; Zhang et al. 2023; Luo et al. 2022)。然而,这些模型存在一定局限性:首先,大多数领域专用模型仅针对医学语料库进行训练,缺乏医学之外多样化知识领域的接触,可能阻碍模型执行推理或上下文理解的能力;其次,这些模型的规模有限,且主要基于 BERT 架构设计,因此在零样本学习下对广泛下游任务的实用性受到限制。本研究旨在通过向通用大语言模型注入医学知识并进行医学专用指令微调,从而解决这两项局限性。
Problem Formulation
问题描述
In this paper, our goal is to systematically investigate the procedure for steering a pre-trained foundational language model to the knowledge-intense domain, i.e., medicine. The training process can be divided into two stages: first, a datacentric knowledge injection stage, that aims to enrich the language model with fundamental medical knowledge; second, a medical-specific instruction tuning stage, that tailors the model to align with clinical use cases.
本文旨在系统研究如何将预训练的基础大语言模型引导至知识密集型领域(如医学)。训练过程分为两个阶段:首先是以数据为中心的知识注入阶段,旨在用基础医学知识丰富语言模型;其次是医学专用指令微调阶段,使模型适配临床用例。
At training stage, assuming the text input as a sequence of tokens, e.g., $\bar{\mathcal{U}}={u_ {1},\bar{u_ {2}},\bar{. }..,u_ {N}}$ , where each $u_ {i}$ is a text token and $N$ is the total sequence length, the training objective is to minimize auto-regressive loss, with the major difference on whether to compute loss on the entire sequence or only sub-sequence, as detailed in the following.
在训练阶段,假设文本输入为一个token序列,例如 $\bar{\mathcal{U}}={u_ {1},\bar{u_ {2}},\bar{. }..,u_ {N}}$ ,其中每个 $u_ {i}$ 是一个文本token, $N$ 是序列总长度。训练目标是最小化自回归损失,主要区别在于计算损失时是针对整个序列还是子序列,具体如下所述。
Data-centric Knowledge Injection. For the knowledge injection step, we simply minimize the default autoregressive loss, all free-form texts on medical knowledge can be used, for the model to accumulate sufficient medicalspecific knowledge contexts, formulated as
以数据为中心的知识注入。在知识注入步骤中,我们仅最小化默认的自回归损失,所有关于医学知识的自由文本均可用于让模型积累足够的医学特定知识上下文,其公式表达为
$$
L(\Phi)=-\sum\log\Phi(u_ {i}|u_ {<i}).
$$
$$
L(\Phi)=-\sum\log\Phi(u_ {i}|u_ {<i}).
$$
where $u_ {<i}$ indicates the tokens appear before index $i$ and $\Phi$ denotes our model.
其中 $u_ {<i}$ 表示索引 $i$ 之前出现的 token,$\Phi$ 代表我们的模型。
Medical-specific Instruction Tuning. At this stage, the token sequence is further split into instruction $\mathcal{T}$ , and response $\mathcal{R}$ , the former is to mimic user’s query, thus the loss is ignored at training time, denoted as:
医疗专用指令微调。在此阶段,token序列被进一步拆分为指令$\mathcal{T}$和响应$\mathcal{R}$,前者用于模拟用户查询,因此在训练时忽略其损失,表示为:
$$
L(\Phi)=-\sum_ {u_ {i}\in\mathcal{R}}\log\Phi(u_ {i}|u_ {<i},\mathcal{T}).
$$
$$
L(\Phi)=-\sum_ {u_ {i}\in\mathcal{R}}\log\Phi(u_ {i}|u_ {<i},\mathcal{T}).
$$
At inference time, the common use case is a conversation, where the user normally provides the question as instruction $\mathcal{T}$ , and the output of the model serves as the answer.
在推理阶段,常见的使用场景是对话交互,用户通常以指令$\mathcal{T}$形式提出问题,模型输出则作为回答。
Dataset Construction
数据集构建
To support our two-stage training, namely data-centric knowledge injection, and medical-specific instruction tuning for alignment, we herein detail the procedure for constructing the high-quality language datasets.
为支持我们的两阶段训练(即数据为中心的知识注入和医疗专用指令微调以实现对齐),本文详细介绍了构建高质量语言数据集的流程。
Dataset-I: Fundamental Medical Knowledge
Dataset-I: 基础医学知识
To steer a general-purpose foundational language model for medical scenario, we propose to first conduct data-centric knowledge injection, that aims to expose the model with medical-related terminologies and definitions. We primarily focus on two key data sources, namely, biomedical papers and textbooks.
为了引导通用基础语言模型适应医疗场景,我们提出首先进行以数据为中心的知识注入,旨在让模型接触医学术语和定义。我们主要关注两个关键数据源:生物医学论文和教科书。
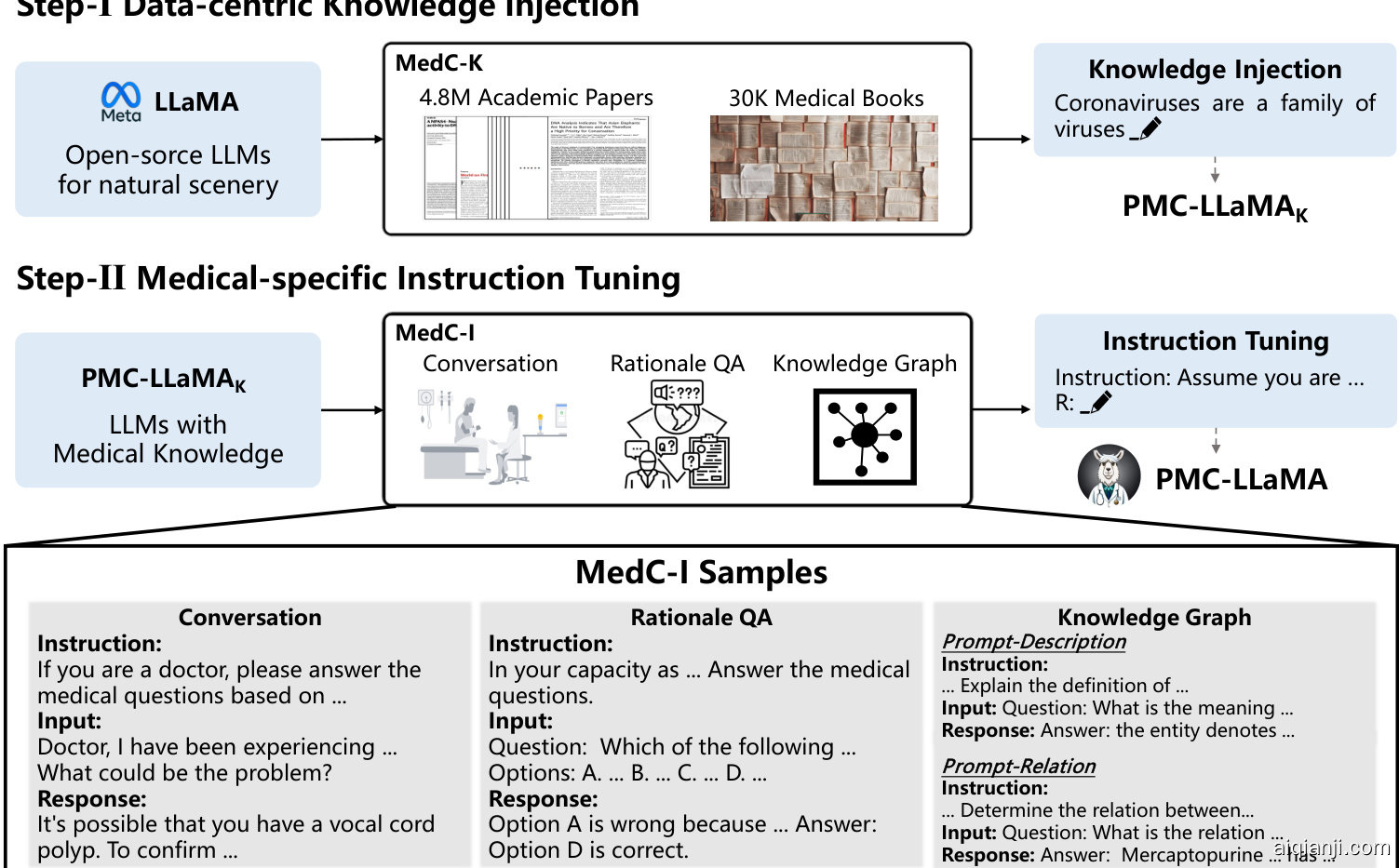

Figure 2: The training pipeline of PMC-LLaMA. Our training flow can be separated into two parts, i.e., data-centric knowledge injection and medical-specific instruction tuning. In knowledge injection, we collect 4.8M biomedical academic papers and 30K medical books for further injecting knowledge into LLaMA. In the instruction tuning stage, we mainly consider three aspects, medical conversation, medical rationale question-answering, and knowledge graph, containing 202M tokens in total.
图 2: PMC-LLaMA 的训练流程。我们的训练流程可分为两部分:以数据为中心的知识注入和医疗专用指令微调。在知识注入阶段,我们收集了480万篇生物医学学术论文和3万本医学书籍,用于向LLaMA进一步注入知识。在指令微调阶段,我们主要考虑三个方面:医疗对话、医学原理问答和知识图谱,总计包含2.02亿个token。
Papers. As a valuable knowledge resource, academic papers naturally contains high-quality, cutting-edge medical knowledge. We start with the S2ORC (Lo et al. 2020) Datasets with 81.1M English-language academic papers, and pick out those biomedical related papers depending on whether having corresponding PubMed Central (PMC) IDs. As a result, there are around 4.8M biomedical papers left, totaling over 75B tokens.
论文。作为宝贵的知识资源,学术论文天然包含高质量、前沿的医学知识。我们从包含8110万篇英文论文的S2ORC数据集 (Lo et al. 2020) 出发,根据是否具有PubMed Central (PMC) ID筛选出生物医学相关论文。最终保留约480万篇生物医学论文,总计超过750亿token。
Books. We collect 30K textbooks sourced from various outlets, for example, the open-library, university library, and reputable publishers, covering a wide range of medical specialties as shown in Fig. 3. For preprocessing, we first extract the text content from the book PDF, then carry out data cleaning via de-duplication and content filtering. Specifically, we eliminate extraneous elements such as URLs, author lists, superfluous information, document contents, references, and citations. Additionally, we have also removed any references to images and tables within the paragraphs, for example, ‘Fig. 1’. After this thorough cleaning process, there are approximately 4B tokens left.
图书。我们收集了来自开放图书馆、大学图书馆和知名出版商等不同渠道的3万本教科书,涵盖广泛的医学专业领域,如图3所示。在预处理阶段,我们首先从图书PDF中提取文本内容,随后通过去重和内容过滤进行数据清洗。具体而言,我们移除了URL、作者列表、冗余信息、文档目录、参考文献和引用等无关内容,并删除了段落中对图表的所有引用(例如"图1")。经过彻底清洗后,剩余约40亿token。
Combination. The two corpora encompass distinct types of medical knowledge, while papers predominantly capture cutting-edge insights, books capture more fundamental medical knowledge, which is more crucial for pre-trained general-purpose language models. Hence, when blending these two datasets for knowledge injection training, we use a ratio of 15:4:1 at each training batch, by that we mean to emphasize “book” tokens more. Specifically, we sample more tokens from books, ensuring they occupy 15 parts per batch and sample tokens from “papers” less so that they occupy 4 parts per batch. For the remaining 1 occupation, we sample from a general language corpus, RedPajama-Data (Computer 2023) to form a complete batch. This mainly aims to avert catastrophic forgetting of previously acquired general text knowledge after extensive knowledge injection on largescale medical-specific data.
组合。这两个语料库涵盖不同类型的医学知识,其中论文主要捕捉前沿见解,而书籍则包含更基础的医学知识,这对预训练的通用语言模型更为关键。因此,在混合这两个数据集进行知识注入训练时,我们在每个训练批次采用15:4:1的比例,即更强调"书籍"token。具体而言,我们从书籍中采样更多token,确保它们每批次占15份,同时减少从"论文"中采样的token,使其每批次占4份。剩余的1份则从通用语言语料库RedPajama-Data (Computer 2023)中采样,以构成完整批次。这主要是为了避免在大规模医学专用数据上进行广泛知识注入后,出现对先前掌握的通用文本知识的灾难性遗忘。
Knowledge Injection Training. Till here, we have constructed a large-scale language dataset of fundamental medical knowledge, termed as MedC-K. With such corpus, we conduct data-centric knowledge injection with autoregressive training, resulting a language model for medicine, named as PMC-LLaMAK, as the largest number of tokens are from PubMed Central academic papers.
知识注入训练。至此,我们已构建了一个基础医学知识的大规模语言数据集,称为MedC-K。基于该语料库,我们采用自回归训练进行以数据为中心的知识注入,最终生成一个医学领域的大语言模型,命名为PMC-LLaMAK(因其绝大部分token源自PubMed Central学术论文)。

Figure 3: Distribution of medical textbooks categories. The box sizes denote the book numbers for different categories.
图 3: 医学教材类别分布。箱体大小表示不同类别的书籍数量。
Dataset-II: Medical Instructions
Dataset-II: 医学指令
Here, we proceed to carry out instruction tuning with the goal of guiding the model to respond to various instructions, by exploiting the medical knowledge embedded in PMC $\mathrm{LLaMA_ {K}}$ model. Generally speaking, our instruction tuning datasets are composed of three main parts, namely, medical consulting conversation, medical rationale QA, and medical knowledge graph prompting.
我们在此进行指令微调,旨在通过利用PMC $\mathrm{LLaMA_ {K}}$ 模型中嵌入的医学知识,引导模型响应各类指令。总体而言,我们的指令微调数据集由三大部分组成:医疗咨询对话、医学原理问答以及医学知识图谱提示。
Medical Conversation. Considering there exists diverse doctor-patient dialogues in daily life, the questions raised by patients are naturally suitable as instructions and doctor responses as ground truth. We start with the data collected by Med-Alpaca (Han, Adams et al. 2023) and ChatDoctor (Yunxiang et al. 2023), and further expand the provided instructions into various synonymous sentences to improve model’s robustness to diverse instructions. Specifically, we use the GPT-4 with the following query prompt:
医疗对话。考虑到日常生活中存在多样化的医患对话,患者提出的问题自然适合作为指令,而医生的回答则作为真实答案。我们从 Med-Alpaca (Han, Adams et al. 2023) 和 ChatDoctor (Yunxiang et al. 2023) 收集的数据入手,进一步将提供的指令扩展为各种同义句,以提高模型对不同指令的鲁棒性。具体而言,我们使用 GPT-4 并输入以下查询提示:
“Rewrite 10 sentences that convey similar meanings to what I’ve stated: {instruction seeds}.”,
重写10个表达与我所述含义相似的句子: {指令种子}。
where {instruction seeds} denotes the provided instruction from ChatDoctor or MedAlpaca, and the query can be repeated until the desired prompt number. At training time, we randomly select one instruction from the instruction base, to simulate the inputs from real users and avoid over-fitting on specific instruction templates.
其中 {instruction seeds} 表示来自 ChatDoctor 或 MedAlpaca 提供的指令,查询可重复执行直至达到所需提示数量。训练时,我们会随机从指令库中选择一条指令,以模拟真实用户输入并避免对特定指令模板过拟合。
Medical Rationale QA. Beyond daily conversations, we also consider equipping our model with reasoning ability with professional medical knowledge. We start with the training sets of the open-source medical multi-choice question-answering datasets, such as USMLE (Jin, Pan et al. 2021), PubMedQA (Jin et al. 2019) and MedMCQA (Pal, Umapathi et al. 2022). Despite the questions in them naturally demanding medical-specific knowledge, most of these datasets only include plain choices, lacking detailed reasoning guidance. To complement such information, we prompt ChatGPT (OpenAI 2023b) for causality analysis. Specifically, given a QA pair, we query ChatGPT to get rationale output (check supplementary for details), and treat the output as an explanation with structured format shown at the bottom of Fig. 2.
医学原理问答。除了日常对话外,我们还考虑为模型配备专业医学知识的推理能力。我们从开源医学多选题数据集的训练集开始,例如USMLE (Jin, Pan et al. 2021)、PubMedQA (Jin et al. 2019)和MedMCQA (Pal, Umapathi et al. 2022)。尽管这些问题天然需要医学专业知识,但大多数数据集仅包含简单选项,缺乏详细的推理指导。为补充此类信息,我们使用ChatGPT (OpenAI 2023b)进行因果分析。具体而言,给定一个问答对,我们查询ChatGPT以获取原理输出(详见补充材料),并将该输出视为结构化格式的解释,如图2底部所示。
Medical Knowledge Graph Prompting. In addition to the aforementioned data, we also consider exploiting medical knowledge graphs UMLS (Lindberg, Humphreys, and McCray 1993), to align with clinicians’ experience. Specifically, to link the medical terminologies with their respective knowledge description or corresponding relationships, we construct QA pairs to translate the common knowledge graph. There are two main types contained in medical knowledge graph, i.e., entity descriptions and entity relationships. We add two different prompts for them as shown at the bottom of Fig. 2, that demands the model to output descriptions for a certain entity or predict the relationship between two entities.
医学知识图谱提示。除上述数据外,我们还考虑利用医学知识图谱UMLS (Lindberg, Humphreys, and McCray 1993) 来与临床医生的经验对齐。具体而言,为将医学术语与其知识描述或对应关系建立关联,我们构建了问答对来转化通用知识图谱。医学知识图谱主要包含两种类型,即实体描述和实体关系。如图2底部所示,我们为它们添加了两种不同的提示,要求模型输出特定实体的描述或预测两个实体间的关系。
Medical-spcific Instruction Tuning. By combining the above three parts together, we form a large-scale, highquality, medical-specific instruction tuning dataset, MedCI, consisting 202M tokens. We further tune PMC-LLaMA $\mathrm{K}$ on it, resulting in our final model – PMC-LLaMA.
医疗领域专用指令微调。通过整合上述三部分内容,我们构建了一个大规模、高质量的医疗领域专用指令微调数据集MedCI,包含2.02亿token。我们在此基础上对PMC-LLaMA $\mathrm{K}$ 进行微调,最终得到我们的目标模型——PMC-LLaMA。
Experiment
实验
Training Details
训练细节
We start by carrying out knowledge injection on open-source LLaMA model, optimizing an auto-regressive loss. Specifically, at training time, the max context length is set as 2048, with a batch size to be 3200, and the model is trained with AdamW optimizer (Loshchilov and Hutter 2017) with a learning rate 2e-5. We adopt the Fully Sharded Data Parallel (FSDP) acceleration strategy, bf16 (Brain Floating Point) data format, and gradient check pointing (Chen et al. 2016). Since we sample more tokens from books in each batch, the model will finish seeing all book tokens earlier. Thus, we here define 1 epoch for seeing all book tokens instead of seeing all mixed tokens. The model is trained with knowledge injection for 5 epochs with 32 A100 GPUs. Then we carry out medical-specific instruction tuning on MedC-I, for 3 epochs with 256 batch sizewith 8 A100 GPUs. Note that, at instruction tuning stage, each epoch refers to looping through all sequences.
我们首先在开源LLaMA模型上进行知识注入,优化自回归损失。具体而言,训练时设置最大上下文长度为2048,批次大小为3200,采用AdamW优化器(Loshchilov and Hutter 2017)进行训练,学习率为2e-5。我们采用全分片数据并行(FSDP)加速策略、bf16(脑浮点)数据格式和梯度检查点(Chen et al. 2016)。由于每个批次从书籍中采样更多token,模型会更早遍历完所有书籍token。因此,我们定义1个epoch为遍历全部书籍token而非混合token。模型使用32块A100 GPU进行5个epoch的知识注入训练。随后在MedC-I数据集上进行医疗专用指令微调,使用8块A100 GPU以256批次大小训练3个epoch。需注意,在指令微调阶段,每个epoch指遍历所有数据序列一次。
Benchmarks
基准测试
In the literature, the primary method for measuring the ability of medical language models is based on multiple-choice question answering, which uses accuracy as the main metric. Following the convention, we adopt three prominent medical question-answering (QA) benchmarks for evaluation.
在文献中,衡量医学语言模型能力的主要方法是基于选择题回答,以准确率作为核心指标。按照惯例,我们采用三个权威的医学问答基准进行评估。
• PubMedQA (Jin et al. 2019) is a biomedical QA dataset collected from PubMed abstracts. The task of PubMedQA is to answer research questions with yes/no/- maybe, which can be considered as the multiple-choice question. It is split into three subsets: 1k manually labeled pairs (PQA-L), $61.2\mathrm{k}$ unlabeled pairs (PQA-U), and 211.3k artificially generated pairs (PQA-A). Following former works (Diao, Pan et al. 2023), we view PQAA as the train set, PQA-L as the test set, and discard the PQA-U parts.
• PubMedQA (Jin et al. 2019) 是一个从PubMed摘要中收集的生物医学问答数据集。该任务要求用"是/否/可能"回答研究问题,可视为多选题。数据集分为三个子集:1k人工标注样本(PQA-L)、$61.2\mathrm{k}$未标注样本(PQA-U)和211.3k人工生成样本(PQA-A)。遵循先前研究(Diao, Pan et al. 2023),我们将PQA-A作为训练集,PQA-L作为测试集,并舍弃PQA-U部分。
• MedMCQA (Pal, Umapathi et al. 2022) is a dataset of multiple choice questions, that are sourced from mock exams and past exams of two Indian medical school entrance exams called AIIMS and NEET-PG (Pal, Umapathi et al. 2022). The train split contains 182,822 questions, and the test split contains 4183 questions. Each question has 4 choices.
• MedMCQA (Pal, Umapathi等, 2022) 是一个多选题数据集,题目来源于印度两所医学院入学考试AIIMS和NEET-PG的模拟试题与历年真题 (Pal, Umapathi等, 2022)。训练集包含182,822道题目,测试集包含4,183道题目。每道题设有4个选项。
• USMLE (Jin, Pan et al. 2021) is a dataset of multiple choice questions (4 choices per question), based on the United States Medical License Exams. The dataset is collected from the professional medical board exams, covering three languages: English, simplified Chinese, and traditional Chinese, containing 12,724, 34,251, and 14,123 questions respectively. Here, we use the English parts and split it into 10,178 questions for training, 1273 for validation, and 1273 for testing, following the official splits.
• USMLE (Jin, Pan et al. 2021) 是一个基于美国医学执照考试的选择题数据集 (每道题4个选项)。该数据集采集自专业医学委员会考试,涵盖英语、简体中文和繁体中文三种语言,分别包含12,724、34,251和14,123道题目。本文采用官方划分的英语部分数据,其中10,178道题用于训练,1,273道用于验证,1,273道用于测试。
Baseline Models
基线模型
LLaMA (Touvron et al. 2023a). LLaMA is the most widely-used open-source language model, it has been trained on a large text corpus with only auto-regressive learning, i.e., no instruction tuning is involved.
LLaMA (Touvron et al. 2023a)。LLaMA是使用最广泛的开源大语言模型,它仅通过自回归学习在大型文本语料库上进行训练,即未涉及指令微调。
LLaMA-2 (Touvron et al. 2023b). LLaMA-2 is the improved version of LLaMA that has been further tuned with instructions. Its largest version (70B) is reported to be the best on natural scenery among the open-source LLMs.
LLaMA-2 (Touvron et al. 2023b)。LLaMA-2是LLaMA的改进版本,经过指令进一步调优。其最大版本(70B)被报告为开源大语言模型中自然场景表现最佳者。
ChatGPT (OpenAI 2023b). ChatGPT is a commercial model released by OpenAI in November, 2022, that has shown remarkable performance on a wide range of NLP tasks in various domains, including medicine. Note that, since the exact details of ChatGPT are confidential, we follow the general presumption that ChatGPT is roughly the same as GPT-3 in model sizes (175B) (Kung et al. 2022).
ChatGPT (OpenAI 2023b)。ChatGPT 是 OpenAI 于 2022 年 11 月发布的商用模型,在包括医学在内的多个领域 NLP 任务中展现出卓越性能。需注意的是,由于 ChatGPT 的具体细节属于机密信息,我们遵循普遍假设,即其模型规模 (175B) 与 GPT-3 大致相当 (Kung et al. 2022)。
Med-Alpaca (Han, Adams et al. 2023). Med-Alpaca is a model further fine-tuned on Alpaca (Taori et al. 2023) using medical instruction data. They focus on the task of assisting medical dialogues and question-answering.
Med-Alpaca (Han, Adams et al. 2023)。Med-Alpaca是基于Alpaca (Taori et al. 2023) 使用医学指令数据进一步微调的模型,其核心目标是辅助医学对话和问答任务。
Chat-Doctor (Yunxiang et al. 2023). Chat-Doctor is a language model aiming for health assistants, that is designed to provide users with medical information, advice, and guidance. For training, it has leveraged the dialogue-based instruction tuning data.
Chat-Doctor (Yunxiang et al. 2023)。Chat-Doctor是一款面向健康助手的语言模型,旨在为用户提供医疗信息、建议和指导。其训练采用了基于对话的指令微调数据。
Evaluation Settings
评估设置
In this section, we describe the evaluating detail to compare the above language models on the QA benchmarks.
在本节中,我们将详细描述如何在问答基准测试上比较上述语言模型的评估细节。
Note that, we do not claim the presented comparison to be completely fair, as a number of training details, for example, data, architecture remain undisclosed for the commercial model. Therefore, we only treat these baseline models for reference, and more focused on presenting our procedure for building on a powerful language model for medicine.
需要注意的是,我们并不声称所呈现的比较完全公平,因为商业模型的许多训练细节(例如数据、架构)仍未公开。因此,我们仅将这些基线模型作为参考,更侧重于展示我们基于强大医学语言模型的构建流程。
Our evaluation settings can be divided into two types: task-specific fine-tuning evaluation and zero-shot instruction evaluation.
我们的评估设置可分为两种类型:任务特定微调评估和零样本指令评估。
Task-specific Fine-tuning Evaluation. In this evaluation setting, we use the combination of three QA training sets to further fine-tune a language model and then evaluate it. For models without instruction tuning, for example, LLaMA and PMC-LLaMAK, we adopt this evaluation setting by default.
任务特定微调评估。在此评估设置中,我们使用三个问答训练集的组合进一步微调语言模型并进行评估。对于未经指令微调的模型(例如LLaMA和PMC-LLaMAK),默认采用此评估设置。
Zero-shot Instruction Evaluation. In this evaluation setting, we directly test the model by giving a medical QA instruction, e.g., “Make a choice based on the question and options.”, without doing any task-specific fine-tuning. Most models are evaluated in this setting, i.e., LLaMA-2, MedAlpaca, Chat-Doctor, ChatGPT, and our own PMC-LLaMA.
零样本指令评估。在此评估设置中,我们直接通过给出医疗问答指令(例如"根据问题和选项做出选择")来测试模型,无需进行任何任务特定的微调。大多数模型在此设置下进行评估,包括 LLaMA-2、MedAlpaca、Chat-Doctor、ChatGPT 以及我们自己的 PMC-LLaMA。
Results
结果
In this section, we will introduce the experimental results. First, we conduct thorough ablation study on medical QA benchmarks, to demonstrate the effectiveness of the different components in our training procedure. Then we show the comparison with different SOTA methods. Lastly, we present qualitative cases studies.
在本节中,我们将介绍实验结果。首先,我们在医疗问答基准上进行了全面的消融研究,以证明训练流程中不同组件的有效性。接着展示了与不同SOTA (state-of-the-art) 方法的对比结果。最后呈现了定性案例分析。
Ablation Study
消融实验
As shown in Tab. 1, we systematically study the different design choices on various medical QA benchmarks, for example, effect of the model scale, data-centric knowledge injection, and medical-specific instruction tuning.
如表 1 所示,我们系统地研究了不同设计选择在各种医疗问答基准上的表现,例如模型规模的影响、以数据为中心的知识注入以及针对医疗领域的指令微调。
Model scale. The scaling law (Kaplan et al. 2020) can also be observed in the medical corpus, for example, as shown in the table, when switching the model size from 7B to 13B, performance on all benchmarks have been improved. This phenomenon holds for both baseline LLaMA model and PMC-LLa $\mathbf{MA}_ {\mathrm{K}}$ , which has further trained with fundamental medical knowledge.
模型规模。缩放定律 (Kaplan et al. 2020) 在医学语料库中同样适用,例如表中所示,当模型规模从 7B 提升至 13B 时,所有基准测试性能均得到提升。这一现象在基线 LLaMA 模型和经过基础医学知识进一步训练的 PMC-LLa $\mathbf{MA}_ {\mathrm{K}}$ 上均成立。
Data-centric knowledge injection. Compared with baseline 7B LLaMA model, integrating biomedical papers brings a performance gain from $44.54%$ to $44.70%$ and $48.51%$ to $50.54%$ on MedQA and MedMCQA respectively. While af-ter adding books for training, the performance is improved significantly, i.e., obtaining $1.02%$ , $2.94%$ , and $1.\bar{2}%$ on MedQA, MedMCQA and PubMedQA respectively. Both observations have shown the importance of injecting fundamental medical knowledge.
以数据为中心的知识注入。相比基线7B LLaMA模型,整合生物医学论文使MedQA和MedMCQA上的性能分别从$44.54%$提升至$44.70%$、$48.51%$提升至$50.54%$。而加入书籍训练后性能显著提升,在MedQA、MedMCQA和PubMedQA上分别获得$1.02%$、$2.94%$和$1.\bar{2}%$的改进。两项观察均证明了注入基础医学知识的重要性。
Medical-specific instruction tuning. We start instruction tuning with only rationale QA data. In this cases, since only QA task is considered, the difference from task-specific finetuning only lies on whether to give rationale sentence as supervision signal. We observe that simply incorporating rationale cases can lead to enhance QA results compared to task-specific fine-tuning on plain choice data, showcasing an improvement of $1.17%$ on the MedQA dataset.
医学专用指令调优。我们仅从原理问答(QA)数据开始指令调优。在这种情况下,由于只考虑QA任务,与任务特定微调的区别仅在于是否提供原理句作为监督信号。我们观察到,相较于仅使用选择题数据的任务特定微调,简单引入原理案例就能提升QA结果,在MedQA数据集上显示出1.17%的改进。
Table 1: Ablation study on QA benchmarks. ACC scores are reported in the table. Note that for the models without ability to follow instruction, we task-specific fine-tune them on the combination of the three downstream training sets to get the number.
| Method | ModelSize | Knowledge Injection | Instruction Tuning e Conversation Knowledge Graph | MedQAMedMCQAPubMedQA | |||||
| Papers | Books | Rationale | |||||||
| Baseline (LLaMA) Baseline (LLaMA) | 7B | × × | × | × × | × × | × | 44.54 45.48 | 48.51 51.42 | 73.40 76.40 |
| PMC-LLaMAK | 13B | ||||||||
| 7B | √ √ | √ | × × | × × | 44.70 | 50.54 | 69.50 | ||
| 7B 13B | √ | √ | × | × | × | 45.56 48.15 | 51.45 54.15 | 74.60 77.10 | |
| PMC-LLaMA | √ | √ | √ | × | |||||
| 13B 13B | √ | √ | √ | √ | × | 49.32 54.43 | 54.56 55.77 | 77.20 77.00 | |
| 13B | √ | √ | √ | √ | √ | 56.36 | 56.04 | 77.90 | |
表 1: QA基准消融研究。表中报告的是ACC分数。请注意,对于不具备指令跟随能力的模型,我们在三个下游训练集的组合上对它们进行了特定任务的微调以获取数据。
| 方法 | 模型大小 | 知识注入 | 指令调优与对话知识图谱 | MedQA | MedMCQA | PubMedQA |
|---|---|---|---|---|---|---|
| 论文 | 书籍 | 原理 | ||||
| Baseline (LLaMA) | 7B | × | × | × | × | × |
| Baseline (LLaMA) | 7B | × | × | × | × | × |
| PMC-LLaMAK | 13B | |||||
| PMC-LLaMAK | 7B | √ | √ | × | × | |
| PMC-LLaMAK | 7B | √ | √ | × | × | × |
| PMC-LLaMAK | 13B | √ | √ | × | × | × |
| PMC-LLaMA | 13B | √ | √ | √ | × | |
| PMC-LLaMA | 13B | √ | √ | √ | √ | × |
| PMC-LLaMA | 13B | √ | √ | √ | √ | × |
| PMC-LLaMA | 13B | √ | √ | √ | √ | √ |
Table 2: Evaluation on QA Benchmarks. ACC scores are reported. Average refers to the average of the three datasets.
| Methods | ModelSize | MedQA | MedMCQA | PubMedQA | Average |
| Human (pass) | 50.0 | 60.0 | |||
| Human (expert) | 87.0 | 90.0 | 78.0 | 85.0 | |
| ChatGPT(OpenAI2023b) | 175B | 57.0 | 44.0 | 63.9 | 54.97 |
| LLaMA-2 (Touvron et al.2023b) | 13B | 42.73 | 37.41 | 68.0 | 49.40 |
| LLaMA-2(Touvron et al.2023b) | 70B | 43.68 | 35.02 | 74.3 | 51.00 |
| Med-Alpaca (Han,Adams et al. 2023) | 13B | 30.85 | 31.13 | 53.2 | 38.38 |
| Chat-Doctor (Yunxiang et al. 2023) | 7B | 33.93 | 31.10 | 54.3 | 39.78 |
| PMC-LLaMA | 13B | 56.36 | 56.04 | 77.9 | 64.43 |
表 2: QA基准测试评估。报告ACC分数。平均值指三个数据集的平均值。
| 方法 | 模型规模 | MedQA | MedMCQA | PubMedQA | 平均值 |
|---|---|---|---|---|---|
| Human (pass) | 50.0 | 60.0 | |||
| Human (expert) | 87.0 | 90.0 | 78.0 | 85.0 | |
| ChatGPT (OpenAI2023b) | 175B | 57.0 | 44.0 | 63.9 | 54.97 |
| LLaMA-2 (Touvron et al.2023b) | 13B | 42.73 | 37.41 | 68.0 | 49.40 |
| LLaMA-2 (Touvron et al.2023b) | 70B | 43.68 | 35.02 | 74.3 | 51.00 |
| Med-Alpaca (Han, Adams et al. 2023) | 13B | 30.85 | 31.13 | 53.2 | 38.38 |
| Chat-Doctor (Yunxiang et al. 2023) | 7B | 33.93 | 31.10 | 54.3 | 39.78 |
| PMC-LLaMA | 13B | 56.36 | 56.04 | 77.9 | 64.43 |
Furthermore, integrating conversations with rationale QA for instruction tuning can produce substantial enhancements, with performance boosts from $49.32%$ to $54.43%$ on MedQA. This demonstrates the pivotal role played by the diversity of question types during the instruction tuning stage, as all involved questions will be limited on medical choice tests without conversation. In addition, the incorporation of a knowledge graph introduces a further improvement of $1.93%$ on the MedQA dataset, demonstrating theimportance of using explicit instructions to emphasize the key medical concepts.
此外,将对话与原理问答结合用于指令微调能带来显著提升,MedQA数据集上的性能从$49.32%$提升至$54.43%$。这表明指令微调阶段问题类型的多样性至关重要,因为若仅包含医学选择题测试而缺乏对话,所有涉及的问题都会受到局限。另外,引入知识图谱使MedQA数据集上的性能进一步提高了$1.93%$,证明通过显式指令强调关键医学概念的重要性。
Comparison with Baselines
与基线方法对比
In Tab. 2, we conduct a comparative analysis of our model against SOTA baseline models on three QA benchmark datasets for evaluation. We also show a qualitative case study to demonstrate the conversation and rationale ability.
在表2中,我们对三个QA基准数据集上的SOTA基线模型进行了对比分析以评估我们的模型。我们还展示了一个定性案例研究来演示对话和推理能力。
Medical QA Ability. While comparing with other large language models on medical QA benchmarks, PMCLLaMA achieves superior results on most of them, improving the average accuracy from $54.97%$ to $64.43%$ , even surpassing the powerful ChatGPT, despite containing significantly fewer parameters.
医疗问答能力。在医学问答基准测试中与其他大语言模型相比,PMCLLaMA在大多数测试中取得了更优的结果,将平均准确率从$54.97%$提升至$64.43%$,尽管参数数量显著更少,甚至超越了强大的ChatGPT。
Zero-shot Case Study. In Fig. 4, we show qualitative examples with the zero-shot prediction from PMC-LLaMA and ChatGPT to verify the quality of prediction, covering patient-physician conversation and rationale QA. The query in Fig. 6a is raised online after our data collection, thus, none of the models have seen this the question at training time. Based on the patient’s description, both PMC-LLaMA and ChatGPT recognize the symptom of recurrent UTIs (urinary tract infections), while PMC-LLaMA proposes a sensitivity test as the specific advice, rather than the general suggestion (investigate the underlying causes) given by ChatGPT. Fig. 6b shows a QA case of microbiology. As can be seen, PMC-LLaMA not only produces the accurate answer, but also briefly analyzes the wrong options, forming a more comprehensive rationale. Another case that focuses on pharmacology knowledge is illustrated in Fig. 6c. Both PMCLLaMA and ChatGPT have shown to properly understand Rifampin’s efficacy and mechanism of side effects.
零样本案例研究。在图4中,我们展示了PMC-LLaMA和ChatGPT的零样本预测定性示例,以验证预测质量,涵盖医患对话和原理问答。图6a中的查询是在我们数据收集后在线提出的,因此所有模型在训练时都未见过该问题。根据患者描述,PMC-LLaMA和ChatGPT都识别出复发性尿路感染(UTI)症状,但PMC-LLaMA提出了敏感性测试作为具体建议,而非ChatGPT给出的通用建议(调查潜在原因)。图6b展示了一个微生物学QA案例。可以看出,PMC-LLaMA不仅给出准确答案,还简要分析了错误选项,形成了更全面的原理说明。图6c展示了另一个聚焦药理学知识的案例,PMC-LLaMA和ChatGPT都正确理解了利福平的药效及副作用机制。
Conclusion
结论
In this paper, we have systematically investigated the procedure for building up a medical-specific large language model based on an open-source large language model, including data-centric knowledge injection and medical-specific instruction tuning. As a result, our proposed PMC-LLaMA is the first, open-source medical-specific language model, that demonstrates superior performance on various medical benchmarks, surpassing ChatGPT and LLaMA-2 with much fewer parameters.
在本文中,我们系统研究了基于开源大语言模型构建医疗领域专用大语言模型的方法,包括以数据为中心的知识注入和医疗领域指令微调。最终提出的PMC-LLaMA成为首个开源的医疗领域专用语言模型,在多项医疗基准测试中展现出超越ChatGPT和LLaMA-2的卓越性能,且参数量显著减少。
Prompt
提示
Prompt
提示
Prompt
提示词
Hello, I am a young and healthy teenage girl, but I suffer from many severe UTIs (urinary tract infections).
你好,我是一名年轻健康的少女,但饱受严重尿路感染 (UTI) 困扰。
I get an average of about eight infections annually over the past four years. I follow all of the conventional methods of hygiene, including vitamin C intake, using cotton underwear, etc. I drink about a gallon of water daily and exercise. This problem has spread to my kidneys the initial time and has never escalated to that state. I developed allergies to all antibiotics, so I am unsure how to treat them at this point. Also, I constantly have cloudy urine and lower back pain.
过去四年里,我平均每年感染约八次。我遵循所有常规卫生方法,包括摄入维生素C、穿棉质内衣等。每天喝约一加仑水并坚持锻炼。这个问题首次发作时就扩散到肾脏,但此后从未恶化到那种程度。我对所有抗生素都产生了过敏反应,因此目前不确定该如何治疗。此外,我还持续出现尿液浑浊和腰痛症状。
Please help.
请帮助。
PMC LLaMA
PMC LLaMA
I have read your query and understand your concern. You seem to be having chronic UTI.
我已阅读您的咨询并理解您的担忧。您似乎患有慢性尿路感染 (UTI)。
You need to get your urine culture and sensitivity test done to know the exact bacteria causing the infection and the antibiotics to which they are sensitive to. Based on the report, you need to take appropriate antibiotics for appropriate duration. Cloudy urine and lower back pain are due to the infection.
你需要进行尿液培养和药敏试验,以确定引起感染的具体细菌及其敏感抗生素。根据报告结果,你需要服用合适的抗生素并确保足够疗程。尿液浑浊和腰痛是由感染引起的。
ChatGPT
ChatGPT
I'm not a doctor, but here are some points you might consider discussing with a healthcare professional:
我不是医生,但以下几点你可以考虑与医疗专业人员讨论:
A microbiologist is studying the emergence of a virulent strain of the virus. After a detailed study of the virus and its life cycle, he proposes a theory: Initially, a host cell is co-infected with 2 viruses from the same virus family. Within the host cell, concomitant production of various genome segments from both viruses occurs. Ultimately, the different genome segments from the viruses are packaged into a unique and novel virus particle. The newly formed virus particle is both stable and viable and is a new strain from the virus family that caused the outbreak of infection. Which of the following viruses is capable of undergoing the abovementioned process?
一位微生物学家正在研究一种高致病性病毒株的出现。经过对病毒及其生命周期的详细研究后,他提出一个理论:最初,宿主细胞同时感染了来自同一病毒家族的两种病毒。在宿主细胞内,两种病毒的各种基因组片段会伴随产生。最终,来自不同病毒的基因组片段被包装成一个独特的新型病毒颗粒。这种新形成的病毒颗粒既稳定又具有生存能力,是引发感染爆发的病毒家族中的新变种。下列哪种病毒能够经历上述过程?
PMC LLaMA
PMC LLaMA
The correct answer is: C. Rotavirus
正确答案是:C. 轮状病毒 (Rotavirus)
A 26-year-old woman presents to a gynecologist after a missed period. After performing a complete physical examination and a screening test for pregnancy, her gynecologist informs her that she is pregnant. She is very surprised as she has been taking oral contraceptives regularly. When the gynecologist asks her about the consumption of any other medications, she mentions that she was placed on treatment for pulmonary tuberculosis (TB) 2 months ago. Her current anti-TB regimen includes rifampin, isoniazid, pyr azin amide, and ethambutol.
一名26岁女性因月经推迟就诊妇科。经全面体检和妊娠筛查测试后,妇科医生告知其已怀孕。患者对此非常惊讶,因她一直在规律服用口服避孕药。当医生询问是否服用其他药物时,她提到两个月前开始接受肺结核 (TB) 治疗,当前抗结核方案包含利福平、异烟肼、吡嗪酰胺和乙胺丁醇。
Which of the following mechanisms best explains the failure of oral contraceptives in this patient?
以下哪种机制最能解释该患者口服避孕药失败的原因?
The correct answer is: C. Rotavirus The description provided corresponds to the process of reassortment or genetic reassortment, which is commonly seen in