SELF-PROMPTING POLYP SEGMENTATION IN COLON OS COPY USING HYBRIDYOLO-SAM 2 MODEL
基于HYBRIDYOLO-SAM 2模型的结肠镜息肉自提示分割方法
ABSTRACT
摘要
Early diagnosis and treatment of polyps during colon os copy are essential for reducing the incidence and mortality of Colorectal Cancer (CRC). However, the variability in polyp characteristics and the presence of artifacts in colon os copy images and videos pose significant challenges for accurate and efficient polyp detection and segmentation. This paper presents a novel approach to polyp segmentation by integrating the Segment Anything Model (SAM 2) with the YOLOv8 model. Our method leverages YOLOv8’s bounding box predictions to autonomously generate input prompts for SAM 2, thereby reducing the need for manual annotations. We conducted exhaustive tests on five benchmark colon os copy image datasets and two colon os copy video datasets, demonstrating that our method exceeds state-of-the-art models in both image and video segmentation tasks. Notably, our approach achieves high segmentation accuracy using only bounding box annotations, significantly reducing annotation time and effort. This advancement holds promise for enhancing the efficiency and s cal ability of polyp detection in clinical settings https://github.com/sajjad-sh33/YOLO_SAM2.
结肠镜检查中息肉的早期诊断和治疗对于降低结直肠癌(CRC)的发病率和死亡率至关重要。然而,息肉特征的多样性以及结肠镜图像和视频中伪影的存在,为准确高效的息肉检测和分割带来了重大挑战。本文提出了一种新颖的息肉分割方法,通过将Segment Anything Model(SAM 2)与YOLOv8模型相结合。我们的方法利用YOLOv8的边界框预测自主生成SAM 2的输入提示,从而减少人工标注的需求。我们在五个结肠镜图像基准数据集和两个结肠镜视频数据集上进行了详尽测试,结果表明我们的方法在图像和视频分割任务中均超越了现有最优模型。值得注意的是,我们的方法仅使用边界框标注就能实现高分割精度,显著减少了标注时间和工作量。这一进展有望提高临床环境中息肉检测的效率和可扩展性https://github.com/sajjad-sh33/YOLO_SAM2。
Index Terms— Colorectal Cancer, Polyp Segmentation, ComputerAided Diagnosis, YOLOv8, Segment Anything.
关键词— 结直肠癌、息肉分割、计算机辅助诊断(ComputerAided Diagnosis)、YOLOv8、Segment Anything
1. INTRODUCTION
1. 引言
Colorectal Cancer (CRC) is one of the leading causes of cancerrelated deaths worldwide, with millions of new cases diagnosed each year [1]. Early detection and removal of polyps during colon os copy significantly reduces the incidence and mortality of CRC. However, the accuracy and efficiency of polyp detection and segmentation remain challenging due to the variability in polyp size, shape, and appearance, as well as the presence of various artifacts and noise in colon os copy images and videos.
结直肠癌 (CRC) 是全球癌症相关死亡的主要原因之一,每年有数百万新病例被确诊 [1]。在结肠镜检查期间早期发现并切除息肉可显著降低结直肠癌的发病率和死亡率。然而,由于息肉大小、形状和外观的多样性,以及结肠镜图像和视频中存在的各种伪影和噪声,息肉检测和分割的准确性与效率仍然具有挑战性。
Computer-Aided Diagnosis (CAD) systems for colon os copy have shown significant potential in enhancing annotation efficiency and reducing the time required for diagnosis. The advent of deep learning has led to the development of numerous neural networks tailored for medical image segmentation [2]. Most current polyp segmentation algorithms rely on variations of the UNet [3] architecture to segment polyps [4]. Additionally, some approaches utilize Res2Net [5] as the backbone and incorporate various regularization techniques to improve segmentation accuracy [6]. Recently, attention-based methods have also been introduced to further enhance performance [7].
计算机辅助诊断 (CAD) 系统在结肠镜检查中展现出显著潜力,可提升标注效率并缩短诊断所需时间。深度学习的发展催生了众多专用于医学图像分割的神经网络 [2]。目前大多数息肉分割算法基于UNet [3] 架构的变体来实现息肉分割 [4]。部分方法采用Res2Net [5] 作为主干网络,并结合多种正则化技术来提高分割精度 [6]。近年来,基于注意力机制的方法也被引入以进一步提升性能 [7]。
Despite these advancements, the process of annotating medical images remains labor-intensive and costly, as it typically requires medical expertise. This challenge has sparked interest in transfer learning, which applies knowledge from large-scale natural image datasets to specific medical imaging tasks. Recent breakthroughs in foundational models, particularly the Segment Anything Model (SAM) [8], have shown exceptional performance in generating highquality object masks from various input prompts. SAM’s success in multiple computer vision benchmarks has gained significant attention for its application in medical image segmentation [9], including polyp image segmentation [10, 11]. The introduction of SAM 2 [12] has further improved real-time segmentation, enabling the processing of entire video sequences based on single-frame annotations. This advancement reduces user interaction time and enhances overall performance, making SAM 2 a valuable tool in medical diagnostics and other applications [13, 14].
尽管取得了这些进展,医学图像的标注过程仍然需要大量人力且成本高昂,因为这通常需要医学专业知识。这一挑战激发了人们对迁移学习的兴趣,即将大规模自然图像数据集中的知识应用于特定医学影像任务。基础模型(尤其是Segment Anything Model (SAM) [8])的最新突破,在根据各种输入提示生成高质量对象掩模方面展现出卓越性能。SAM在多项计算机视觉基准测试中的成功,使其在医学图像分割[9](包括息肉图像分割[10, 11])中的应用获得了广泛关注。SAM 2[12]的推出进一步提升了实时分割能力,支持基于单帧标注处理整个视频序列。这一进展减少了用户交互时间并提升了整体性能,使SAM 2成为医学诊断等应用中的宝贵工具[13, 14]。
Despite the strong zero-shot capabilities of SAM 2 for segmenting medical images, it typically requires input prompts provided by human experts. This reliance on manual input limits the efficiency and s cal ability of the segmentation process. To address this limitation, we propose a self-prompting segmentation model that autonomously generates input prompts by integrating the YOLOv8 [15] model’s pre-trained capabilities. By combining YOLO’s bounding box predictions with SAM 2’s segmentation capabilities, our method aims to enhance the accuracy and efficiency of polyp segmentation in colon os copy images and videos. In this work, we utilize only bounding box data for training our model to perform the segmentation task. This approach significantly reduces annotation time compared to previous methods that required detailed ground truth segmentation masks for training. By leveraging bounding box annotations, our model can achieve high segmentation accuracy with less manual effort, making it more practical for large-scale applications. This approach not only tackles variability in polyp features but also reduces the computational load for large-scale segmentation tasks.
尽管SAM 2在医学图像分割中展现出强大的零样本能力,但它通常需要人类专家提供输入提示。这种对人工输入的依赖限制了分割过程的效率和可扩展性。为解决这一局限,我们提出了一种自提示分割模型,通过整合YOLOv8 [15]模型的预训练能力自主生成输入提示。通过结合YOLO的边界框预测与SAM 2的分割能力,我们的方法旨在提升结肠镜图像和视频中息肉分割的准确性与效率。本研究中,我们仅使用边界框数据训练模型执行分割任务。与先前需要详细真实分割掩模进行训练的方法相比,该方法显著减少了标注时间。通过利用边界框标注,我们的模型能以更少的人工投入实现高分割精度,使其更适合大规模应用。该方法不仅解决了息肉特征的多样性问题,还降低了大规模分割任务的计算负担。
2. METHODOLOGY
2. 方法
Our proposed model integrates two state-of-the-art algorithms, YOLO and SAM 2, to effectively detect and segment polyps in colon os copy images and videos. The process begins with the YOLO model, which identifies potential polyps and places bounding boxes around them. These bounding boxes are then employed as input prompts for the SAM 2 model, which performs precise segmentation of the polyps based on the provided coordinates, Fig. 1.
我们提出的模型整合了两种先进算法 YOLO 和 SAM 2,能有效检测并分割结肠镜图像和视频中的息肉。该流程首先通过 YOLO 模型识别潜在息肉并生成边界框,随后将这些边界框作为输入提示传递给 SAM 2 模型,后者根据提供的坐标对息肉进行精确分割 (图 1)。
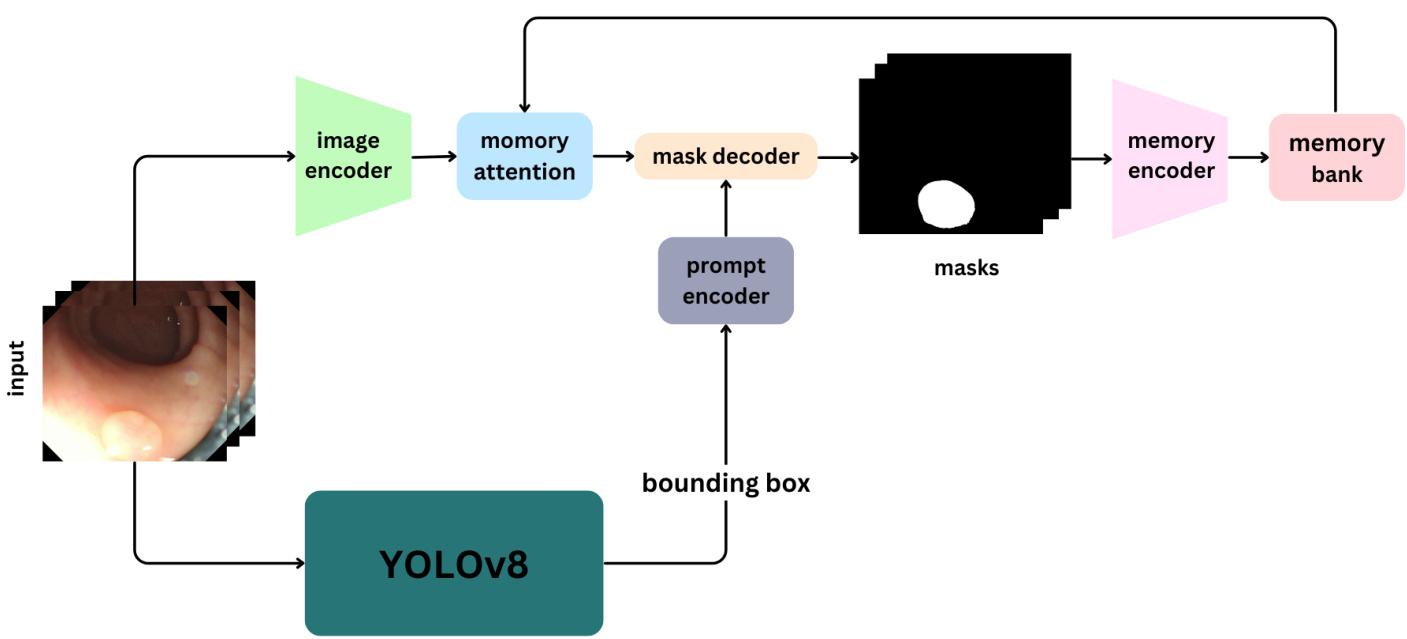
Fig. 1: Architecture of the Self-Prompting YOLO-SAM 2 Model for Polyp Segmentation.
图 1: 用于息肉分割的自提示YOLO-SAM 2模型架构。
A pre-trained YOLOv8 model is employed for its superior speed and accuracy in real-time object detection tasks. YOLOv8 processes the colon os copy images through a Convolutional Neural Network (CNN), extracting essential features and predicting bounding boxes around potential polyps. The box coordinates of these bounding boxes are then passed to the SAM 2 model as prompts.
采用预训练的YOLOv8模型进行实时目标检测任务,因其兼具卓越速度与精度。YOLOv8通过卷积神经网络(CNN)处理结肠镜图像,提取关键特征并预测潜在息肉周围的边界框。随后将这些边界框的坐标作为提示信息传递给SAM 2模型。
The SAM 2 model, known for its lightweight nature and high accuracy, refines the detection results provided by YOLOv8. It uses bounding box coordinates to perform detailed segmentation, delineating the exact boundaries of the polyps. The SAM 2 model architecture consists of several key components: Image Encoder: Utilizes a transformer-based architecture to extract high-level features from both images and video frames. This component is responsible for understanding visual content at each timestep. Prompt Encoder: Processes user-provided prompts to guide the segmentation task. This allows SAM 2 to adapt to user input and target specific objects within a scene. Memory Mechanism: Includes a memory encoder, memory bank, and memory attention module. These components collectively store and utilize information from past frames, enabling the model to maintain consistent object tracking over time. Mask Decoder: Produces the final segmentation masks based on the encoded image features and prompts.
SAM 2模型以其轻量级和高精度著称,能够优化YOLOv8提供的检测结果。它利用边界框坐标进行精细分割,精确勾勒出息肉的实际边界。SAM 2模型架构包含以下关键组件:
图像编码器 (Image Encoder):采用基于Transformer的架构,从图像和视频帧中提取高级特征。该组件负责理解每个时间步的视觉内容。
提示编码器 (Prompt Encoder):处理用户提供的提示以指导分割任务,使SAM 2能够适应用户输入并定位场景中的特定目标。
记忆机制 (Memory Mechanism):包含记忆编码器、记忆库和记忆注意力模块。这些组件共同存储并利用历史帧信息,使模型能够保持持续的目标追踪。
掩码解码器 (Mask Decoder):根据编码后的图像特征和提示生成最终的分割掩码。
Due to the lightweight nature of both models, it is still possible to use their combination as a real-time segmentation model to segment polyp videos. During training, only the YOLOv8 model would be fine-tuned based on the bounding box dataset, while the SAM 2 weights are frozen and not fine-tuned.
由于两种模型都具有轻量级特性,它们的组合仍可作为实时分割模型用于分割息肉视频。训练过程中,仅基于边界框数据集对YOLOv8模型进行微调,而SAM 2的权重保持冻结状态不进行微调。
It is important to emphasize that in this study, the YOLOv8 model is dedicated solely to generating bounding boxes rather than segmentation masks. Our approach focuses on utilizing only bounding box data to train the overall segmentation model, leveraging the zero-shot capabilities of SAM 2 to minimize the need for extensive data annotation. Additionally, our findings indicate that SAM 2 provides superior segmentation performance than YOLOv8.
需要强调的是,在本研究中,YOLOv8模型仅用于生成边界框而非分割掩码。我们的方法专注于仅利用边界框数据来训练整体分割模型,借助SAM 2的零样本能力以最大限度减少大量数据标注的需求。此外,我们的研究结果表明,SAM 2的分割性能优于YOLOv8。
3. EXPERIMENTS AND RESULTS
3. 实验与结果
3.1. Datasets
3.1. 数据集
In order to assess the performance of the SAM 2 model, we carried out comparative experiments utilizing five well-known benchmark colon os copy image datasets, along with two additional video colon os copy datasets. The following sections provide detailed descriptions of each dataset. 1) Kvasir-SEG [24]: Curated by the Vestre Viken Health Trust in Norway, this dataset includes 1,000 polyp images and their corresponding ground truth from colon os copy video sequences. 2) CVC-ClinicDB [25]: Created in collaboration with the Hospital Clinic of Barcelona, Spain, it contains 612 images from colon os copy examination videos, originating from 29 different sequences. 3) CVC-ColonDB [25]: This dataset comprises 380 polyp images, each with its corresponding ground truth, captured at a resolution of $500\times570$ pixels from 15 distinct videos. 4) ETIS [26]: It includes 196 polyp images, each captured at a resolution of $966\times1225$ pixels, aiding research in polyp detection and analysis. 5) CVC-300 [27]: Comprising 60 polyp images, each captured at a resolution of $500\times574$ pixels. 6) PolypGen [28]: A comprehensive dataset for polyp detection and segmentation, including 1,537 polyp images, 4,275 negative frames, and 2,225 positive video sequences, collected from six medical centers across Europe and Africa. 7) SUN-SEG [29, 30]: This dataset includes 158,690 frames from 113 colon os copy videos, with detailed annotations for each frame.
为了评估SAM 2模型的性能,我们利用五个知名的基准结肠镜图像数据集和两个额外的结肠镜视频数据集进行了对比实验。以下对各数据集进行详细说明。
- Kvasir-SEG [24]: 由挪威Vestre Viken Health Trust整理,包含来自结肠镜视频序列的1,000张息肉图像及其对应标注真值。
- CVC-ClinicDB [25]: 与西班牙巴塞罗那Hospital Clinic合作创建,包含来自29个不同序列的结肠镜检查视频中的612张图像。
- CVC-ColonDB [25]: 该数据集包含380张息肉图像,每张图像均附带标注真值,从15个不同视频中以$500\times570$像素分辨率采集。
- ETIS [26]: 包含196张息肉图像,每张图像以$966\times1225$像素分辨率采集,用于辅助息肉检测与分析研究。
- CVC-300 [27]: 包含60张息肉图像,每张图像以$500\times574$像素分辨率采集。
- PolypGen [28]: 一个综合性的息肉检测与分割数据集,包含1,537张息肉图像、4,275张阴性帧和2,225段阳性视频序列,数据采集自欧洲和非洲的六个医疗中心。
- SUN-SEG [29, 30]: 该数据集包含113段结肠镜视频中的158,690帧,每帧均有详细标注。
3.2. Implementation Details
3.2. 实现细节
As mentioned earlier, the SAM model remains frozen, and only the YOLOv8 model is trained. After conducting experiments, we chose the YOLOv8 medium version due to its superior performance compared to the large and small versions. Despite having 25 million parameters, it effectively handles real-time video segmentation when combined with the SAM 2 model. Additionally, we use the SAM 2 large model, which contains 224.4 million parameters. Despite its larger size, it maintains real-time performance, processing approximately 44 frames per second. This makes it suitable for applications requiring high accuracy and speed, such as video analysis and interactive segmentation tasks. For training, $80%$ of the dataset is used, with the remaining $20%$ reserved for evaluation. We set the batch size to 64 and the input image size to 680. We implemented our model using A100 (40 GB) GPU.
如前所述,SAM模型保持冻结状态,仅训练YOLOv8模型。经过实验验证,我们选择了YOLOv8中型版本,因其性能优于大型和小型版本。尽管该版本拥有2500万参数,但与SAM 2模型结合后仍能高效处理实时视频分割任务。此外,我们采用包含2.244亿参数的SAM 2大型模型,其在大规模参数下仍保持实时性能,每秒可处理约44帧,非常适合视频分析和交互式分割等高精度、高速度应用场景。训练时使用数据集的80%,剩余20%用于评估,批次大小设为64,输入图像尺寸为680。模型在A100 (40GB) GPU上完成实现。
Table 1: A quantitative comparison of five benchmark datasets with state-of-the-art (SOTA) methods is presented. The best performance i highlighted in bold.
表 1: 五个基准数据集与当前最优 (SOTA) 方法的定量对比。最佳性能以粗体标出。
| 方法 | CVC-ClinicDB mIoU mDice | Kvasir-SEG mIoU mDice | CVC-ColonDB mIoU mDice | ETIS mIoU mDice | CVC-300 mIoU mDice |
|---|---|---|---|---|---|
| UNet [3] | 0.755 0.823 | 0.746 0.818 | 0.436 0.504 | 0.335 0.398 | 0.627 0.710 |
| UNet++ [16] | 0.729 0.794 | 0.744 0.821 | 0.408 0.482 | 0.344 0.401 | 0.624 0.707 |
| MSEG [17] | 0.864 0.909 | 0.839 0.897 | 0.666 0.735 | 0.630 0.700 | 0.804 0.874 |
| SANet [7] | 0.859 0.916 | 0.847 0.904 | 0.669 0.752 | 0.654 0.750 | 0.815 0.888 |
| MSNet [18] | 0.869 0.918 | 0.849 0.905 | 0.668 0.747 | 0.650 0.720 | 0.796 0.862 |
| SSFormer [19] | 0.855 0.906 | 0.864 0.917 | 0.721 0.802 | 0.720 0.796 | 0.827 0.895 |
| CFA-Net [6] | 0.883 0.933 | 0.861 0.915 | 0.665 0.743 | 0.655 0.732 | 0.827 0.893 |
| Polyp-PVT [20] | 0.905 0.948 | 0.864 0.917 | 0.727 0.808 | 0.706 0.787 | 0.833 0.900 |
| SAM-Path [21] | 0.644 0.750 | 0.730 0.828 | 0.516 0.632 | 0.442 0.555 | 0.756 0.844 |